
|
Telecom Paris
Dep. Informatique & Réseaux  J-L. Dessalles
← Home pageNovember 2024
J-L. Dessalles
← Home pageNovember 2024 |

 Module Athens TP-09
Emergence in Complex Systems
Module Athens TP-09
Emergence in Complex Systems
| Ada Diaconescu - Associate professor at Telecom-Paris |
| Jean-Louis Dessalles - Associate professor at Telecom-Paris |
| Samuel Reyd - PhD student at Telecom-Paris |

Evolife

Presentation
This software has been developed at Telecom ParisTech by Jean-Louis Dessalles for this course.
Evolife is written in Python. The package contains a documentation, which can also be found there.
Evolife’s latest sources may be downloaded from there.
To execute Evolife on your machine, see ’How to execute Evolife’ in the documentation.Evolife implements various aspects of evolutionary simulation, including:
- Genetic Algorithms
- Darwinian ecology
- Collective phenomena
First contact with Evolife
 Get Evolife and install it,
as explained.
Get Evolife and install it,
as explained.
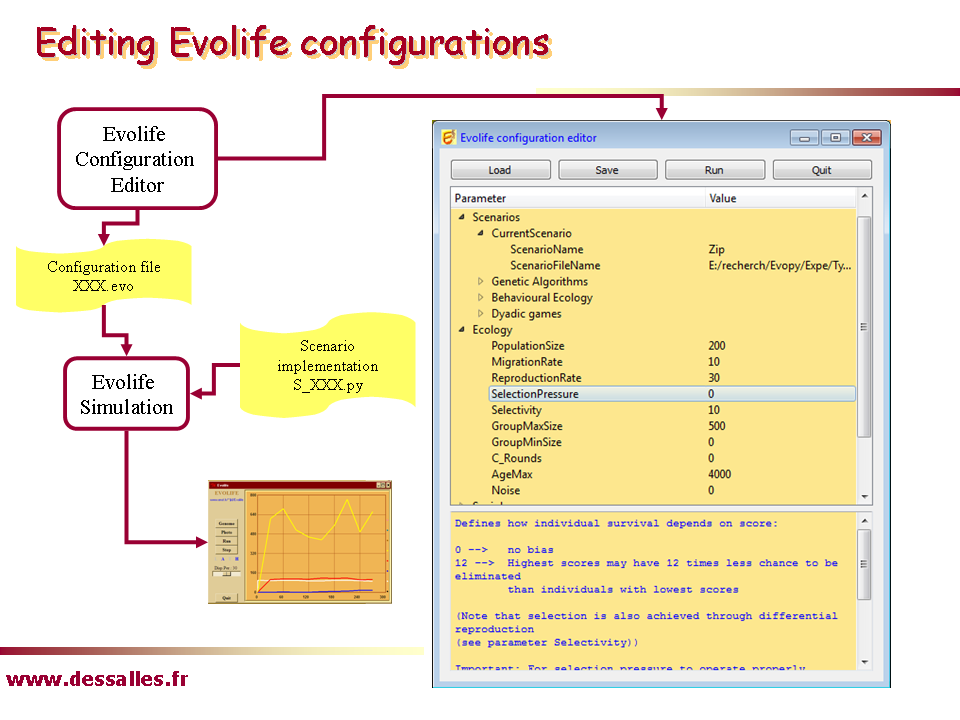
Note: if your are using a unix or Linux machine, don’t forget to run the command "python first.py" once, as explained in the documentation.Run Starter (in the Evolife directory). It opens the Configuration Editor. There are a number of pre-stored configuration files (*.evo files) that can be loaded with the [Load] button. Most of these files are located in the folder Expe. You may click on parameter values in the Configuration Editor to change them (alternatively, you can navigate using arrows and use the [F2] key to edit values). Contextual documentation is given in the bottom panel.Example: Load the SumBits.evo configuration file from Starter. Press [Run] (shortcut Alt-R). The Evolife window should open. Click its [Run] button to run the simulation (this experiment will be studied in the section "Genetic Algorithms").
Buttons
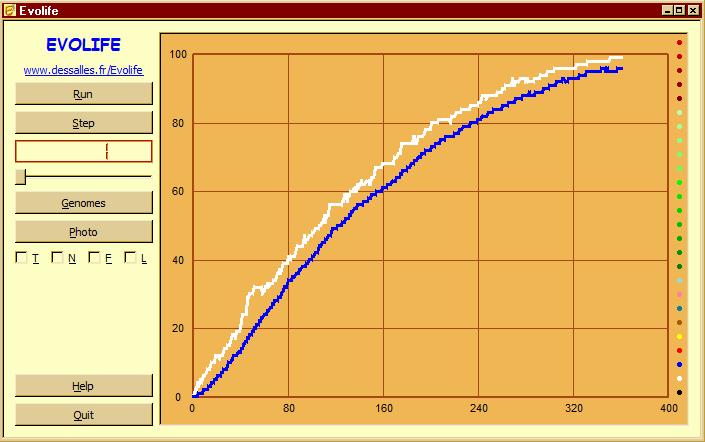
[Genome]: Displays the genomes of the population
- Each horizontal line represents the genome of one individual
- Vertical patterns result from correlations between individuals
- Prints a few data on the console
- Saves a screenshot for each opened window. Screenshots are saved in .png files (format can be changed on demand).
If you enter Film mode (by pressing key ‘v'), photos are taken automatically at each display period. These files can be assembled into animated gifs, e.g. using ImageMagick. - Saves the current plots in ‘Evolife.csv’ (which may be opened with Excel).
This file is also created when exiting Evolife.
[Step]: Runs the simulation stepwise
[T]: Displays trajectories when applicable
[N]: Displays social links (network) when applicable
[F]: Displays agents’ positions on a 2-D field
[L]: Log messages (not implemented)
[Help]: Displays help file
Shortcuts
- <Esc>: quits the active window (and the application if main window is active) (like ‘q')
- <F1>: displays this help file
- a: displays alliances (social links between individuals, sorted by competence) (when applicable)
- c: runs the simulation in continuous mode (like ‘r')
- f: displays a 2-D field on which agents may be located
- g: displays genomes
- h: displays this help file
- l: displays log messages (not implemented)
- m: alternatively brings main window or satellite windows to foreground
- q: quits the active window (and the application if main window is active) (like
) - r: runs the simulation in continuous mode (like ‘c')
- s: runs the simulation one step forward (like
) - t: displays trajectories (when applicable)
- v: enters film mode. One picture per opened window is recorded
- z: shrinks satellite windows (genomes, alliances, labyrinth...) (dezoom)
- -: shrinks satellite windows (genomes, alliances, labyrinth...) (dezoom)
- +: expands satellite windows (genomes, alliances, labyrinth...) (zoom)
(shortcuts also work from satellite windows)Each configuration file XXX.evo corresponds to a scenario, implemented in a file named S_XXX.py located in the Scnenarii directory. Try to locate the S_SumBits.py. Open it in your favourite editor. You can observe that its main class, the class Scenario, inherits Default_Scenario defined in the file Default_Scenario.py. This is the main way you will be using Evolife. A particular scenario is defined by redefining some of the default functions defined in Default_Scenario. For instance, the scenario corresponding to S_SumBits.py redefines functions such as genemap or evaluation.In case of error: Errors may occur, as you will be asked to change the program. Most errors will be displayed in the text window. However, in case of python crash, use a terminal window, go to the Evolife directory and execute Main.py. The python error will be displayed.
| Please express your thoughts, remarks, suggestions to improve Evolife. |
 Back to the main page
Back to the main page